|
chr6_-_128883125
|
4.860
|
NM_002844
NM_001135648
|
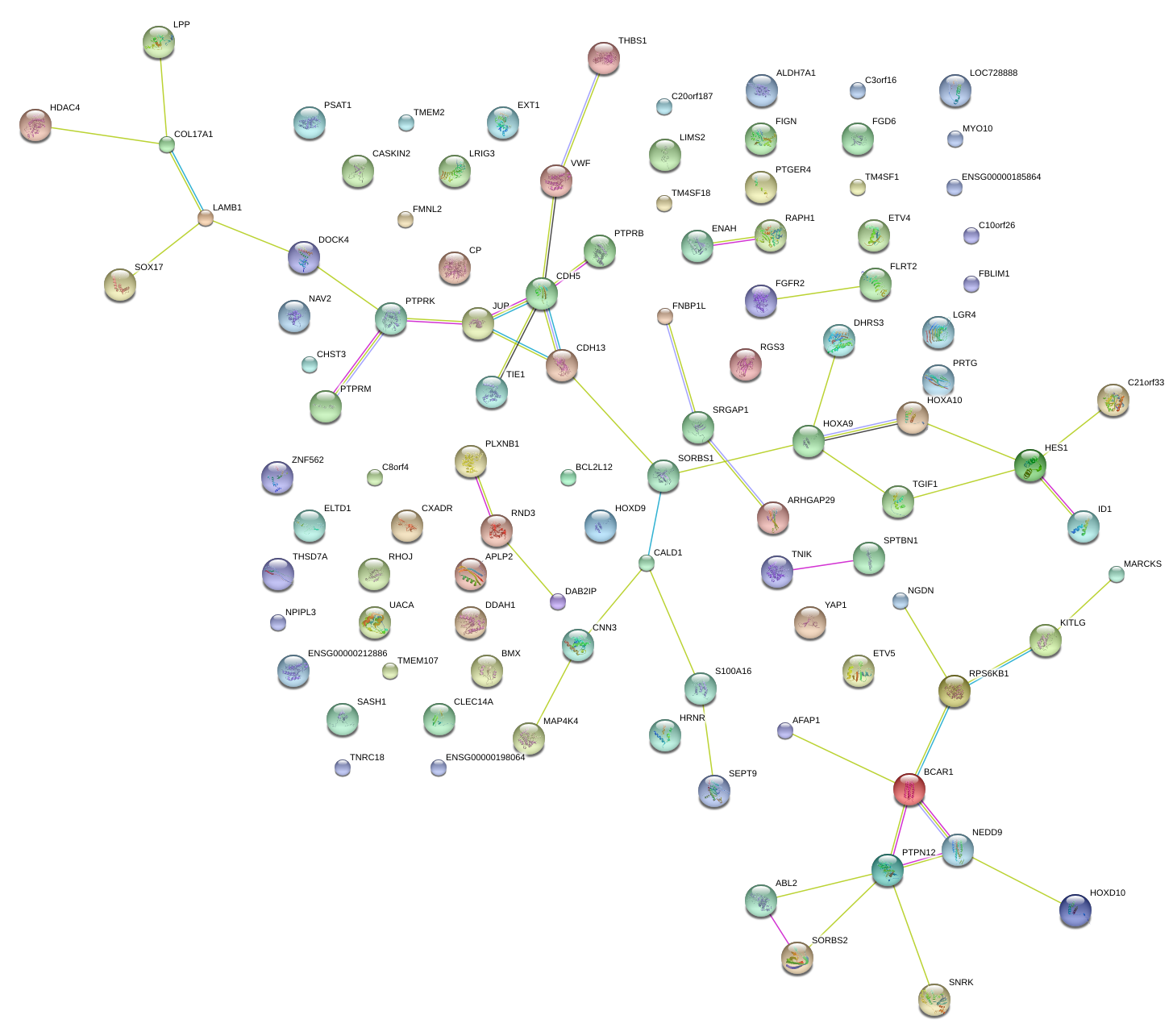
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr2_+_101680594
|
3.051
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_-_27171673
|
2.429
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr12_-_6103945
|
2.407
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr3_+_195336625
|
2.369
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr7_-_27171648
|
2.337
|
|
HOXA9
|
homeobox A9
|
|
chr7_-_27171600
|
2.332
|
|
HOXA9
|
homeobox A9
|
|
chr6_-_11340869
|
2.268
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11340886
|
2.213
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr6_-_11340887
|
2.197
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr3_-_187309442
|
2.016
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr4_-_7991992
|
2.008
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr5_-_16989371
|
1.985
|
NM_012334
|
MYO10
|
myosin X
|
|
chr9_+_123501186
|
1.970
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr6_+_148705421
|
1.957
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr5_-_16989290
|
1.929
|
|
MYO10
|
myosin X
|
|
chr17_-_76985538
|
1.844
|
|
|
|
|
chr1_-_95165037
|
1.800
|
|
CNN3
|
calponin 3, acidic
|
|
chr3_-_189354510
|
1.793
|
|
LOC339929
|
hypothetical LOC339929
|
|
chr21_+_17807167
|
1.762
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr1_-_151848039
|
1.728
|
|
S100A16
|
S100 calcium binding protein A16
|
|
chr10_+_73393998
|
1.721
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr3_-_150422474
|
1.703
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chrX_+_15428820
|
1.655
|
NM_203281
|
BMX
|
BMX non-receptor tyrosine kinase
|
|
chr10_-_97311084
|
1.647
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr17_-_37196463
|
1.627
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr7_-_5429702
|
1.624
|
NM_001080495
|
TNRC18
|
trinucleotide repeat containing 18
|
|
chr9_+_115303527
|
1.614
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr11_+_101488380
|
1.560
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr1_+_15957832
|
1.557
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr3_+_189354167
|
1.540
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr14_+_85066265
|
1.538
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr10_+_104525877
|
1.528
|
NM_017787
|
C10orf26
|
chromosome 10 open reading frame 26
|
|
chr1_-_94475504
|
1.522
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_+_195336631
|
1.499
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr14_+_62740878
|
1.497
|
NM_020663
|
RHOJ
|
ras homolog gene family, member J
|
|
chr16_+_81218142
|
1.474
|
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr14_-_37795290
|
1.468
|
NM_175060
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr6_+_114285209
|
1.467
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr8_+_40130143
|
1.465
|
NM_020130
|
C8orf4
|
chromosome 8 open reading frame 4
|
|
chr11_+_129496792
|
1.449
|
|
APLP2
|
amyloid beta (A4) precursor-like protein 2
|
|
chr7_-_27186400
|
1.402
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr1_-_223907283
|
1.386
|
NM_001008493
NM_018212
|
ENAH
|
enabled homolog (Drosophila)
|
|
chr5_+_40715778
|
1.363
|
NM_000958
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr1_-_85703198
|
1.354
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr5_+_40715799
|
1.340
|
|
PTGER4
|
prostaglandin E receptor 4 (subtype EP4)
|
|
chr16_+_81218074
|
1.339
|
NM_001257
|
CDH13
|
cadherin 13, H-cadherin (heart)
|
|
chr1_+_207668787
|
1.338
|
NM_001104548
|
LOC642587
|
NPC-A-5
|
|
chr14_+_85066207
|
1.332
|
NM_013231
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr8_-_119192985
|
1.329
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr7_+_77005287
|
1.282
|
NM_001131008
NM_001131009
|
PTPN12
|
protein tyrosine phosphatase, non-receptor type 12
|
|
chr20_+_29656744
|
1.281
|
NM_002165
NM_181353
|
ID1
|
inhibitor of DNA binding 1, dominant negative helix-loop-helix protein
|
|
chr14_-_37795001
|
1.279
|
|
CLEC14A
|
C-type lectin domain family 14, member A
|
|
chr2_-_164300758
|
1.259
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr11_+_20005704
|
1.256
|
|
NAV2
|
neuron navigator 2
|
|
chr15_+_37660568
|
1.255
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr2_-_239987557
|
1.254
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr1_-_12599934
|
1.250
|
NM_004753
|
DHRS3
|
dehydrogenase/reductase (SDR family) member 3
|
|
chr1_-_95165089
|
1.249
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr12_+_62524778
|
1.243
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr3_-_150533948
|
1.231
|
NM_001184723
NM_138786
|
TM4SF18
|
transmembrane 4 L six family member 18
|
|
chr2_-_151052391
|
1.215
|
NM_005168
|
RND3
|
Rho family GTPase 3
|
|
chr1_-_177378801
|
1.193
|
NM_001136000
NM_001168239
NM_005158
|
ABL2
|
v-abl Abelson murine leukemia viral oncogene homolog 2
|
|
chr11_-_27450808
|
1.191
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr6_-_72187168
|
1.190
|
|
C6orf155
|
chromosome 6 open reading frame 155
|
|
chr1_-_197173159
|
1.185
|
|
LOC100131234
|
familial acute myelogenous leukemia related factor
|
|
chr8_+_55533047
|
1.175
|
NM_022454
|
SOX17
|
SRY (sex determining region Y)-box 17
|
|
chr7_-_11838260
|
1.164
|
NM_015204
|
THSD7A
|
thrombospondin, type I, domain containing 7A
|
|
chr12_-_57600564
|
1.141
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr1_+_93686154
|
1.131
|
NM_001024948
NM_001164473
NM_017737
|
FNBP1L
|
formin binding protein 1-like
|
|
chr17_+_72795567
|
1.123
|
NM_001113492
|
SEPT9
|
septin 9
|
|
chr4_-_186969041
|
1.120
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr14_+_23008723
|
1.119
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr1_+_43539239
|
1.117
|
NM_005424
|
TIE1
|
tyrosine kinase with immunoglobulin-like and EGF-like domains 1
|
|
chr18_+_3441589
|
1.112
|
NM_170695
NM_173210
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr3_+_43303005
|
1.106
|
NM_001100594
NM_017719
|
SNRK
|
SNF related kinase
|
|
chr16_-_21344158
|
1.105
|
NM_130464
|
NPIPL3
|
nuclear pore complex interacting protein-like 3
|
|
chr15_-_53822468
|
1.102
|
NM_173814
|
PRTG
|
protogenin
|
|
chr15_-_68781662
|
1.082
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr2_-_204108151
|
1.073
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr16_+_64958025
|
1.070
|
NM_001795
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr16_+_64958060
|
1.069
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr2_-_56266408
|
1.052
|
|
LOC100129434
|
hypothetical LOC100129434
|
|
chr1_-_79244948
|
1.048
|
NM_022159
|
ELTD1
|
EGF, latrophilin and seven transmembrane domain containing 1
|
|
chr7_-_107430566
|
1.047
|
NM_002291
|
LAMB1
|
laminin, beta 1
|
|
chr2_-_128138435
|
1.045
|
NM_017980
|
LIMS2
|
LIM and senescent cell antigen-like domains 2
|
|
chr5_-_125958503
|
1.043
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr18_+_7936885
|
1.041
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr2_+_54536780
|
1.041
|
NM_003128
|
SPTBN1
|
spectrin, beta, non-erythrocytic 1
|
|
chr12_-_94135154
|
1.034
|
NM_018351
|
FGD6
|
FYVE, RhoGEF and PH domain containing 6
|
|
chr7_-_111633428
|
1.028
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr10_-_105835525
|
1.020
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr10_-_105835609
|
1.016
|
NM_000494
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr3_-_150993299
|
1.016
|
NM_001144960
|
C3orf16
|
chromosome 3 open reading frame 16
|
|
chr16_-_73842884
|
1.002
|
NM_001170717
NM_014567
|
BCAR1
|
breast cancer anti-estrogen resistance 1
|
|
chr9_+_80101810
|
0.999
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr7_+_134114957
|
0.995
|
|
CALD1
|
caldesmon 1
|
|
chr17_-_71017509
|
0.987
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr7_+_134114912
|
0.985
|
|
CALD1
|
caldesmon 1
|
|
chr3_-_150578023
|
0.973
|
|
TM4SF1
|
transmembrane 4 L six family member 1
|
|
chr12_-_69289889
|
0.972
|
NM_002837
|
PTPRB
|
protein tyrosine phosphatase, receptor type, B
|
|
chr9_-_73573121
|
0.966
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr14_+_23008742
|
0.956
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr12_-_87498225
|
0.950
|
NM_000899
NM_003994
|
KITLG
|
KIT ligand
|
|
chr16_+_64958086
|
0.942
|
|
CDH5
|
cadherin 5, type 2 (vascular endothelium)
|
|
chr2_+_152900076
|
0.939
|
|
FMNL2
|
formin-like 2
|
|
chr10_-_123346148
|
0.934
|
NM_001144915
|
FGFR2
|
fibroblast growth factor receptor 2
|
|
chr2_-_239987629
|
0.929
|
|
HDAC4
|
histone deacetylase 4
|
|
chr3_-_48445719
|
0.926
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr17_-_38979178
|
0.912
|
NM_001079675
|
ETV4
|
ets variant 4
|
|
chr16_+_22432344
|
0.911
|
NM_001135865
|
LOC100132247
NPIPL3
|
nuclear pore complex interacting protein related gene
nuclear pore complex interacting protein-like 3
|
|
chr3_-_101077605
|
0.908
|
NM_014890
|
FILIP1L
|
filamin A interacting protein 1-like
|
|
chr9_+_96807107
|
0.904
|
|
C9orf3
|
chromosome 9 open reading frame 3
|
|
chr4_+_74491272
|
0.903
|
|
ALB
|
albumin
|
|
chr19_+_60543032
|
0.897
|
NM_032701
|
SUV420H2
|
suppressor of variegation 4-20 homolog 2 (Drosophila)
|
|
chr1_-_68470585
|
0.889
|
NM_001002292
NM_001193334
NM_024911
|
WLS
|
wntless homolog (Drosophila)
|
|
chr1_+_167342149
|
0.883
|
|
ATP1B1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr16_-_52877829
|
0.879
|
|
IRX3
|
iroquois homeobox 3
|
|
chr9_+_27099138
|
0.879
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr3_+_138020550
|
0.872
|
NM_001097599
NM_001097600
NM_025246
|
TMEM22
|
transmembrane protein 22
|
|
chr11_-_128567302
|
0.868
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr12_-_51173238
|
0.867
|
NM_005554
|
KRT6A
|
keratin 6A
|
|
chr12_+_94135645
|
0.865
|
NM_017599
|
VEZT
|
vezatin, adherens junctions transmembrane protein
|
|
chr2_-_70329067
|
0.846
|
NM_022037
NM_022173
|
TIA1
|
TIA1 cytotoxic granule-associated RNA binding protein
|
|
chr12_-_57600336
|
0.845
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr20_-_55718982
|
0.842
|
NM_199169
NM_199170
|
PMEPA1
|
prostate transmembrane protein, androgen induced 1
|
|
chr18_+_3439972
|
0.840
|
NM_173209
NM_173208
NM_003244
|
TGIF1
|
TGFB-induced factor homeobox 1
|
|
chr2_+_37425240
|
0.837
|
NM_012413
|
QPCT
|
glutaminyl-peptide cyclotransferase
|
|
chr20_-_13567529
|
0.833
|
NM_017714
|
TASP1
|
taspase, threonine aspartase, 1
|
|
chr6_-_2785682
|
0.832
|
|
SERPINB1
|
serpin peptidase inhibitor, clade B (ovalbumin), member 1
|
|
chr5_+_149960820
|
0.832
|
NM_001166208
|
SYNPO
|
synaptopodin
|
|
chrX_+_67965517
|
0.827
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr4_-_89963177
|
0.819
|
NM_001015045
|
FAM13A
|
family with sequence similarity 13, member A
|
|
chr19_+_50663089
|
0.805
|
NM_001114171
NM_006732
|
FOSB
|
FBJ murine osteosarcoma viral oncogene homolog B
|
|
chr5_-_92942680
|
0.804
|
|
FLJ42709
|
hypothetical LOC441094
|
|
chr7_+_12693729
|
0.798
|
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr12_+_52665196
|
0.793
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr12_-_106011715
|
0.790
|
NM_004075
|
CRY1
|
cryptochrome 1 (photolyase-like)
|
|
chr1_+_67923660
|
0.789
|
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chrX_-_44945025
|
0.785
|
NM_024689
NM_176819
|
CXorf36
|
chromosome X open reading frame 36
|
|
chr10_-_105982052
|
0.779
|
NM_025145
|
WDR96
|
WD repeat domain 96
|
|
chr1_+_67923331
|
0.777
|
NM_001924
|
GADD45A
|
growth arrest and DNA-damage-inducible, alpha
|
|
chr8_+_21980315
|
0.775
|
|
EPB49
|
erythrocyte membrane protein band 4.9 (dematin)
|
|
chr15_-_39973566
|
0.771
|
NM_016642
|
SPTBN5
|
spectrin, beta, non-erythrocytic 5
|
|
chr1_+_176577228
|
0.760
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr17_-_59817456
|
0.760
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_+_24795465
|
0.759
|
|
KIAA1217
|
KIAA1217
|
|
chr1_-_94475840
|
0.757
|
NM_004815
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr10_+_24795446
|
0.756
|
|
KIAA1217
|
KIAA1217
|
|
chr9_-_74757735
|
0.755
|
NM_000689
|
ALDH1A1
|
aldehyde dehydrogenase 1 family, member A1
|
|
chr5_-_50714836
|
0.753
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr17_+_75366524
|
0.748
|
NM_005189
NM_032647
|
CBX2
|
chromobox homolog 2
|
|
chr15_+_68971835
|
0.748
|
NM_001199017
NM_017691
NM_001199018
|
LRRC49
|
leucine rich repeat containing 49
|
|
chr3_+_190990142
|
0.747
|
NM_001114980
NM_001114981
NM_001114982
|
TP63
|
tumor protein p63
|
|
chr6_-_10523424
|
0.740
|
NM_003220
|
TFAP2A
|
transcription factor AP-2 alpha (activating enhancer binding protein 2 alpha)
|
|
chr17_-_59818033
|
0.735
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr2_+_160180092
|
0.732
|
|
LOC643072
|
hypothetical LOC643072
|
|
chr14_+_22375753
|
0.731
|
|
MMP14
|
matrix metallopeptidase 14 (membrane-inserted)
|
|
chr9_-_35101534
|
0.731
|
|
KIAA1539
|
KIAA1539
|
|
chr12_-_108802561
|
0.724
|
NM_016433
|
GLTP
|
glycolipid transfer protein
|
|
chr6_+_30702586
|
0.724
|
NM_024909
NM_001031722
NM_001190724
|
ATAT1
|
alpha tubulin acetyltransferase 1
|
|
chr7_+_116099648
|
0.723
|
NM_000245
NM_001127500
|
MET
|
met proto-oncogene (hepatocyte growth factor receptor)
|
|
chr6_+_43565192
|
0.720
|
NM_001146017
|
TJAP1
|
tight junction associated protein 1 (peripheral)
|
|
chr1_+_144439069
|
0.720
|
NM_002614
|
PDZK1
|
PDZ domain containing 1
|
|
chr1_-_167721723
|
0.716
|
NM_006996
|
SLC19A2
|
solute carrier family 19 (thiamine transporter), member 2
|
|
chr16_+_27468942
|
0.715
|
NM_015202
|
KIAA0556
|
KIAA0556
|
|
chrX_+_70359780
|
0.714
|
NM_000166
|
GJB1
|
gap junction protein, beta 1, 32kDa
|
|
chr1_+_85819004
|
0.713
|
NM_001554
|
CYR61
|
cysteine-rich, angiogenic inducer, 61
|
|
chr1_-_33585848
|
0.708
|
|
|
|
|
chr16_-_52877868
|
0.703
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr11_-_88864061
|
0.700
|
NM_001143836
NM_016931
|
NOX4
|
NADPH oxidase 4
|
|
chr18_+_18003401
|
0.699
|
NM_005257
|
GATA6
|
GATA binding protein 6
|
|
chr4_-_159312973
|
0.698
|
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr4_-_101658094
|
0.690
|
NM_001159694
NM_016242
|
EMCN
|
endomucin
|
|
chr17_-_59817654
|
0.685
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr10_-_33287184
|
0.683
|
NM_002211
|
ITGB1
|
integrin, beta 1 (fibronectin receptor, beta polypeptide, antigen CD29 includes MDF2, MSK12)
|
|
chr1_-_6468600
|
0.681
|
NM_001042665
|
PLEKHG5
|
pleckstrin homology domain containing, family G (with RhoGef domain) member 5
|
|
chr9_-_35101592
|
0.675
|
|
KIAA1539
|
KIAA1539
|
|
chr13_-_40138607
|
0.673
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr5_+_50714996
|
0.668
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr6_-_29752909
|
0.668
|
NM_001109809
|
ZFP57
|
zinc finger protein 57 homolog (mouse)
|
|
chr16_+_464844
|
0.666
|
NM_001142272
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr4_-_110942576
|
0.665
|
|
CFI
|
complement factor I
|
|
chr2_+_62786290
|
0.662
|
NM_001142616
NM_015252
|
EHBP1
|
EH domain binding protein 1
|
|
chr7_+_16652282
|
0.656
|
NM_001159767
NM_014038
|
BZW2
|
basic leucine zipper and W2 domains 2
|
|
chr4_-_110942783
|
0.655
|
NM_000204
|
CFI
|
complement factor I
|
|
chr16_-_30705707
|
0.654
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr7_+_12693435
|
0.653
|
NM_001195396
NM_005738
|
ARL4A
|
ADP-ribosylation factor-like 4A
|
|
chr6_-_80303822
|
0.652
|
NM_001122769
NM_181714
|
LCA5
|
Leber congenital amaurosis 5
|
|
chr18_+_54039818
|
0.652
|
|
NEDD4L
|
neural precursor cell expressed, developmentally down-regulated 4-like
|
|
chrX_-_106846253
|
0.647
|
NM_001015881
|
TSC22D3
|
TSC22 domain family, member 3
|
|
chr20_-_17459973
|
0.645
|
NM_001195
|
BFSP1
|
beaded filament structural protein 1, filensin
|
|
chr17_-_59817575
|
0.644
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr5_+_148186348
|
0.644
|
NM_000024
|
ADRB2
|
adrenergic, beta-2-, receptor, surface
|
|
chr4_-_186969202
|
0.640
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_159313167
|
0.640
|
NM_016613
|
FAM198B
|
family with sequence similarity 198, member B
|
|
chr2_+_159021653
|
0.638
|
|
PKP4
|
plakophilin 4
|
|
chr14_-_54948290
|
0.636
|
NM_014924
|
ATG14
|
ATG14 autophagy related 14 homolog (S. cerevisiae)
|
|
chr12_-_10142779
|
0.635
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr12_+_51730101
|
0.634
|
NM_170754
|
TENC1
|
tensin like C1 domain containing phosphatase (tensin 2)
|